Generalized Expectation Maximization [1]
This technical report describes the statistical method of expectation maximization (EM) for parameter estimation. Several of 1D, 2D, 3D and n-D examples are presented in this document. Applications of the EM method are also demonstrated in the case of mixture modeling using interactive Java applets in 1D (e.g., curve fitting), 2D (e.g., point clustering and classification) and 3D (e.g., brain tissue classification).
Table
of Contents
1
Maximum Likelihood Estimation (MLE)
2
General Expectation Maximization (GEM) Algorithms
Application
1 (Missing Values)
Application
2 (Pattern Recognition)
3 EM
Application: Finding Maximum Likelihood Mixture Densities Parameters via EM
Example
3: 1D Distribution Mixture-Model-Fitting using EM
Example
4: 2D Point Clustering and Classification using EM
Example
5: 3D Brain Tissue Classification using EM and Mixture Modeling
1 Maximum Likelihood Estimation (MLE)
First, let’s
recall the definition of the maximum-likelihood estimation problem. We have a
density function p
(x | Q), that is governed by the set of parameters Q (e.g.,
p might be a Gaussian and Q could be the means (vector) and covariance
(matrix)). We also have a data set of size N, supposedly drawn
from this distribution with density p, i.e., X = {x1,
…, xN}. That is, we assume that these data vectors are
independent and identically distributed (IID) with distribution p . Therefore, the resulting joint density for
the samples is:
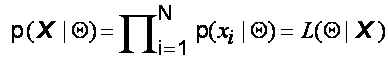
![]() is called the likelihood
function of the parameters (Q)
given the data, or just the likelihood. The likelihood is thought of as a
function of the parameters (Q) where
the data X is fixed (observed). In the maximum likelihood problem, our goal is to
find a parameter vector Q that
maximizes
is called the likelihood
function of the parameters (Q)
given the data, or just the likelihood. The likelihood is thought of as a
function of the parameters (Q) where
the data X is fixed (observed). In the maximum likelihood problem, our goal is to
find a parameter vector Q that
maximizes ![]() . In other words, we
look for Q*
, where
. In other words, we
look for Q*
, where
![]()
Oftentimes we
choose to maximize ![]() instead because it is analytically easier or
computationally appealing.
instead because it is analytically easier or
computationally appealing.
Depending on the
form of p (x | Q)
this problem can be easy or hard. For example, if p (x | Q)
is simply a single Gaussian distribution where the parameter vector Q=(m, s2),
then we can solve the
maximum likelihood problem of determining estimates (MLE) of m & s2
by setting the partial derivatives of ![]() to zero (in fact, this
is how the familiar formulas for the population mean and variance are obtained). For many problems, however, it is not
possible to find such analytical expressions, and we must resort to more
elaborate techniques (e.g., EM technique).
to zero (in fact, this
is how the familiar formulas for the population mean and variance are obtained). For many problems, however, it is not
possible to find such analytical expressions, and we must resort to more
elaborate techniques (e.g., EM technique).
Example 1: Nornal(m,s2)
Suppose {X1, …, Xn} IID N(m, s2),
where m is
unknown. We estimate m by: ![]() .
.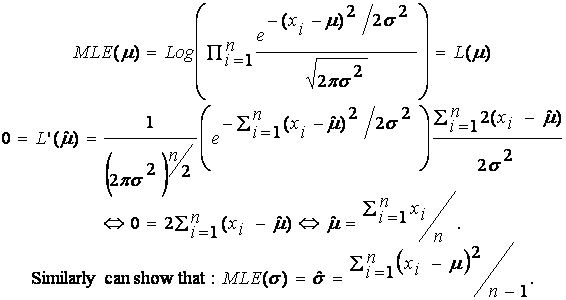
Example 2: Poisson(l)
Suppose {X1, …, XN} IID Poisson(l),
where (the population mean, and standard deviation) l is
unknown. Estimate l by:
![]()

2 General Expectation Maximization (GEM) Algorithms
The EM algorithm
is one such technique, which allows estimating parameter vectors in the cases
when such analytic solutions to the likelihood minimization are difficult or
impossible. The EM algorithm [see references at the end] is a general
method of finding the maximum-likelihood estimates of the parameters of an
underlying distribution from a given data set when the data is incomplete or has
missing values. There are two main applications of the EM algorithm.
Application 1 (Missing Values)
The first application of EM algorithm occurs when the data indeed has missing values, due to problems with or limitations of the observation process.
Application 2 (Pattern Recognition)
The second occurs
when optimizing the likelihood function is analytically intractable, however,
we still need to assume the likelihood function can be simplified by assuming
the existence of and values for additional but missing (or hidden)
parameters. This second application is more common in the computational pattern
recognition community.
As before, we
assume that data X is observed and is generated by some
distribution. We call X the incomplete
data. We assume that a complete data set exists Z=(X;Y) and also assume (or specify) a joint density
function:
![]()
Recall that for each probability measure, P, P(A,B)=P(A|B)P(B)
and hence, in terms of the conditional probability, P(A,B|C) =
P(A|B,C)P(B|C). This joint density often times arises from the marginal
density function ![]() and the assumption of
hidden variables and parameter value guesses (e.g., mixture-densities, this
example is coming up; and Baum-Welch algorithm). In other cases (e.g., missing
data values in samples of a distribution), we must assume a joint relationship
between the missing and observed values.
and the assumption of
hidden variables and parameter value guesses (e.g., mixture-densities, this
example is coming up; and Baum-Welch algorithm). In other cases (e.g., missing
data values in samples of a distribution), we must assume a joint relationship
between the missing and observed values.
With this new
density function, we can define a new likelihood function,
![]() ,
,
called the complete-data likelihood. Note that this
function is in fact a random variable since the missing information Y
is unknown, i.e., random, and presumably governed by an underlying
distribution. That is, we can think of ![]() for some function
for some function ![]() , where X and Q are constant
and Y is a random variable.
The original likelihood
, where X and Q are constant
and Y is a random variable.
The original likelihood ![]() is referred to as the incomplete-data likelihood function.
is referred to as the incomplete-data likelihood function.
The Expectation
Maximization algorithm then proceeds in two steps – expectation followed by its
maximization.
Step 1 of EM (Expectation)
The EM algorithm
needs to first find the expected value of the complete-data log-likelihood ![]() with respect to the unknown data Y given the observed data X and the current parameter estimates Qi, the E-Step. Below we define
the expectation of Q(i-1), the second argument represents the
parameters that we use to evaluate the expectation. The first argument Q simply indicates the parameter (vector) that
ultimately will be optimized in an attempt to maximize the likelihood:
with respect to the unknown data Y given the observed data X and the current parameter estimates Qi, the E-Step. Below we define
the expectation of Q(i-1), the second argument represents the
parameters that we use to evaluate the expectation. The first argument Q simply indicates the parameter (vector) that
ultimately will be optimized in an attempt to maximize the likelihood:
Q(Q, Q(i-1))=E[Log p (X,Y | Q) | X, Q(i-1)]. (1)
Where Q(i-1) are the current parameters estimates that we
used to evaluate the expectation and Q are
the new parameters that we optimize to increase (maximize) Q. Note that the expression above is a conditional
expectation w.r.t. (X & Q(i-1)),
i.e.,
E[ h(Y) |
X=x] :=![]() .
.
The key thing to
understand is that X and Q(i-1) are given constants, Q is a random variable that we wish to
adjust/estimate, and Y is a
random variable governed by the distribution f(Y |X,Q(i-1)). The right side of
equation (1) can therefore be expressed as:
![]() (2)
(2)
The f(Y |X,Q(i-1)) is the marginal
distribution of the unobserved data (Y) and is dependent on: the
observed data X, on the
current parameters Q(i-1), and Y is the space of values y can
take on. In the best-case situations, this marginal distribution is a
simple analytical expression of the assumed parameters Q(i-1), and perhaps
the observed data (X). In the worst-case scenario, this
density might be very hard to obtain. In fact, sometimes the actually used
density is:
f(y
,X | Q(i-1))= f(y | X,Q(i-1)) f(X | Q(i-1)),
but this doesn’t
effect subsequent steps since the extra factor, f(X | Q(i-1)) is not
dependent on Q.
As an analogy,
suppose we have a function h( . ; . ) of two variables. Consider h( q ; Y ) where q is a
constant and Y is a random
variable governed by some distribution fY(y). Then
![]()
is now a
deterministic function that could be maximized if desired, w.r.t. q. The evaluation of this expectation is
called the E-step of the
algorithm. Notice the meaning of the two arguments in the function Q(Q, Q(i-1)). The first argument Q corresponds
to the parameters that ultimately will be optimized in an attempt to maximize
the likelihood. The second argument, Q(i-1), corresponds to the parameters that we use
to evaluate the expectation at each iteration (EàM à EàM à EàM à …).
Step 2 of EM (Maximization)
The second step
(the M-step) of the EM algorithm is to maximize the expectation
we computed in the first step. That is, we iteratively compute:
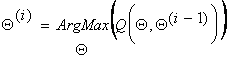
That is we maximize the expectation of the log-likelihood function. These two steps are repeated as necessary. Each iteration is guaranteed to increase the log-likelihood and the algorithm is guaranteed to converge to a local maximum of the likelihood function. There are many theoretical and empirical rate-of-convergence papers (see references below).
A modified form of
the M-step is to, instead of maximizing the rather difficult function Q(Q, Q(i-1)), we find some Q(i) such that Q(Q(i), Q(i-1)) > Q(Q, Q(i-1)). This form of the algorithm is called
Generalized EM (GEM) and is also guaranteed to converge. This
description of the GEM does not yield a direct computer implementation scheme
(it’s not constructive (the coding algorithm is not explicit). This is the way,
however, that the algorithm is presented in its most general form. The details
of the steps required to compute the given quantities are very dependent on the
particular application, so they are not discussed when the algorithm is
presented in this abstract form, but rather detailed for each specific
application.
3 EM Application: Finding Maximum Likelihood Mixture Densities Parameters via EM
The
mixture-density parameter estimation problem is probably one of the most widely
used applications of the EM algorithm in the computational pattern recognition
community. In this case, we assume the following (mixture-density) model:
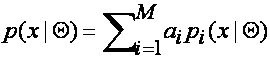
![]()
where the
parameter vector is Q=(a1, …, aM ; q1, …, qM), where the mixture-model weights satisfy ![]() and each pi is
a density function parameterized, in general, by its own parameter vector qi. In other words, we assume we have M component
densities mixed together with M mixing coefficients ai .
and each pi is
a density function parameterized, in general, by its own parameter vector qi. In other words, we assume we have M component
densities mixed together with M mixing coefficients ai .
The
incomplete-data log-likelihood expression for this density from the data X={x1,
…, xN}, N = # Observations,
is given by:
![]()
which is difficult
to optimize because it contains the logarithm function of the sum [if the sum
and the log were interchanged then optimizing the outside logarithm would have
been equivalent to optimizing its argument – the sum – as the log function has
always a positive derivative over its domain (0; ![]() )]. If we consider X as incomplete, however, and
assume the existence of unobserved data items Y={yi}i=1N, whose values inform us which component
density “generated” each data item, the likelihood expression is significantly
simplified. That is, if we assume that
)]. If we consider X as incomplete, however, and
assume the existence of unobserved data items Y={yi}i=1N, whose values inform us which component
density “generated” each data item, the likelihood expression is significantly
simplified. That is, if we assume that ![]() for each
for each ![]() , and yi=k if the ith
sample, xi, was generated by the kth mixture component pk. If we
know the values of Y, the likelihood becomes:
, and yi=k if the ith
sample, xi, was generated by the kth mixture component pk. If we
know the values of Y, the likelihood becomes:
![]()
which, given a
particular form of the component densities, can be optimized using a variety of
techniques. The problem, of course, is that we do not know the values of Y.
If we assume Y is a random
vector, however, we can proceed. We first must derive an expression for the
distribution of the unobserved (missing) data, Y. Let’s first
guess at parameters for the mixture density, i.e., we guess that Qg =(a1g,
…, aMg
; q1g, …, qMg), are the
appropriate parameters for the likelihood L(Qg | X,Y). Given Qg, we can compute pj(xi|qjg) for each i
and j. In addition, the mixing parameters, ai, can
be though of as prior probabilities of each mixture component, that is ai= p(component i). Therefore, using Bayes’s rule – P(Yi|Xi)=P(Xi|Yi)P(Yi)/P(Xi),
we can compute:

and therefore, ![]() , where y = (y1,
…, yN)
is an instance of the unobserved
data independently drawn. When we now look at equation (2), we see that in this
case we have obtained the desired marginal density (of Y), f(y
| X,Q(i-1)), by assuming the existence of the hidden variables and making a guess
at the initial parameters of their distribution. In this case, equation (1)
takes the specific form:
, where y = (y1,
…, yN)
is an instance of the unobserved
data independently drawn. When we now look at equation (2), we see that in this
case we have obtained the desired marginal density (of Y), f(y
| X,Q(i-1)), by assuming the existence of the hidden variables and making a guess
at the initial parameters of their distribution. In this case, equation (1)
takes the specific form:
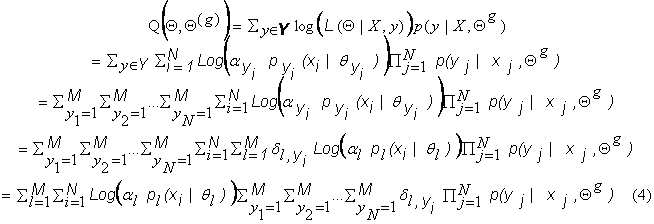
where 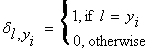 . In this form,
. In this form, ![]() appears computationally challenging, however,
it can be simplified, since for
appears computationally challenging, however,
it can be simplified, since for ![]() ,
,
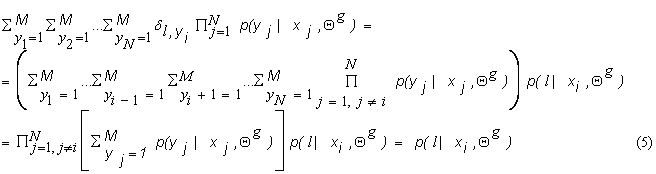
This is because ![]() . Using equation (5), we can write equation (4) as:
. Using equation (5), we can write equation (4) as:
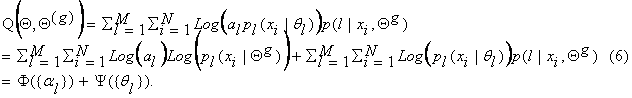
Now, to maximize
this expression, we can maximize independently F and
Y, the terms containing al and ql, since
they are not related. To find the expression for al, which maximizes Q(Q,Q(g)), we introduce the Lagrange multiplier l with the constraint that ![]() , and solve
the following equation:
, and solve
the following equation:
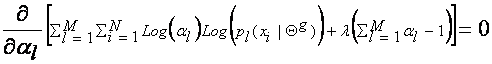
This yields: 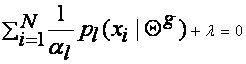
Summing over l we get:
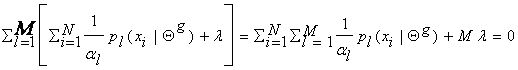
Therefore, l = -N , which yields that 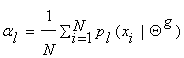 . This is how we determine the mixture parameters {al}, most of the time.
. This is how we determine the mixture parameters {al}, most of the time.
Now let us try to estimate the second part of the
parameter vector Q=(a1,
…, aM ; q1, …, qM), i.e., the distribution specific parameters (q1, …, qM). Clearly, these need to be estimated in a case by case manner, as
different distributions have different number and type of parameters. We
consider again a couple of cases that illustrate the basic strategy for
estimating (q1, …, qM) using EM approaches. For some distributions, it will be possible to
get analytic expressions for ql directly, as functions of all other
variables.
4. Examples
Example 1: Poisson(l)
Suppose that the mixture model in equation (3) involves
Poisson(ll)
distributions, ![]() . Then the
. Then the
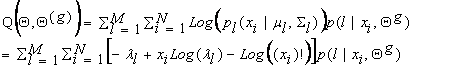
Taking the
derivatives w.r.t. ll and setting these equal to zero yields,

Therefore, we have explicit expressions for
iterative calculation of the estimates of the mixture parameters, Q=(a1, …, aM), and the Poisson distribution parameters, (q1, …, qM)=(l1, …, lM).
Example 2: n-D Gaussian
If we assume d-dimensional
Gaussian component distributions with a mean vector m and covariance matrix S, i.e., q = (m; S) then the
probability density function is
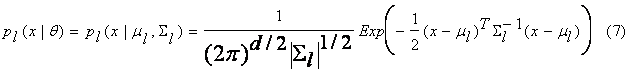
![]()
Then we may derive
the update equations [equations (1), (4), (6)] for this specific distribution,
we need to recall some results from matrix algebra. The trace of a
square matrix tr(A) is
equal to the sum of A’s diagonal elements. In 1D the trace of a
scalar equals that scalar. Also, tr(A + B) = tr(A) + tr(B), and tr(AB) = tr(BA), which
implies that ![]() . Also note that |A| indicates the determinant of the
matrix A, and |A-1|=1/|A|. Differentiation of a
function of a matrix f(A) is accomplished by differentiating with respect to elements of
that matrix. Therefore, we define df(A)/dA to be the matrix with
(i,j)-th entry equal to [df(A)/dai,j], where A=( ai,j).
This definition also applies taking derivatives with respect to a vector.
First, d(xTAx)/dx=(A+AT)x. Second, it can be shown
that when A is a symmetric matrix:
. Also note that |A| indicates the determinant of the
matrix A, and |A-1|=1/|A|. Differentiation of a
function of a matrix f(A) is accomplished by differentiating with respect to elements of
that matrix. Therefore, we define df(A)/dA to be the matrix with
(i,j)-th entry equal to [df(A)/dai,j], where A=( ai,j).
This definition also applies taking derivatives with respect to a vector.
First, d(xTAx)/dx=(A+AT)x. Second, it can be shown
that when A is a symmetric matrix:
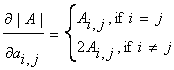
where Ai,j
is the (i,j)-the cofactor of A. Given the above, we see that:
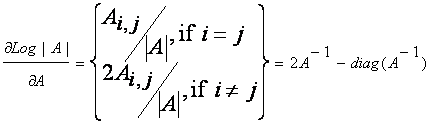
by the definition
of the inverse of a matrix. Finally, it can be shown that dtr(AB)/dA
= B + BT-diag(B).
Now, for the d-dimensional
Gaussian distribution example, if we take a log of equation (7), ignoring any
constant terms (since they disappear after taking derivatives), and
substituting into the right side of equation (6), we get:

Taking the
derivative of equation (8) with respect to ml and setting it equal to zero, we get:
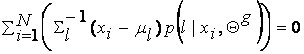
which solving for ml yields:
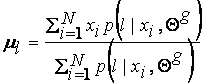
To find Sl, note that we can write equation (8) as:
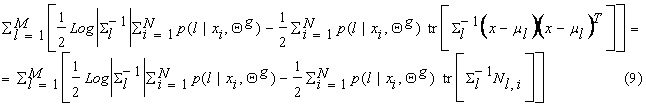
where Nl,i = ![]() . In equation (9)
we now take the derivative with respect to the matrix Sl-1, and we obtain:
. In equation (9)
we now take the derivative with respect to the matrix Sl-1, and we obtain:

where Nl,i = ![]() , Ml,i =
Sl – Nl,i and S =
, Ml,i =
Sl – Nl,i and S = 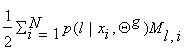 . To find the extreme
values (maxima) of the , equation (9) we set the derivative to zero [equation
(10)], i.e., 2S – diag(S) = 0. This implies that S=0 è
. To find the extreme
values (maxima) of the , equation (9) we set the derivative to zero [equation
(10)], i.e., 2S – diag(S) = 0. This implies that S=0 è ![]() So, we obtain an exact
expression (variance-covariance matrix estimate, 1
So, we obtain an exact
expression (variance-covariance matrix estimate, 1 ![]() l
l ![]() M) for Sl.
M) for Sl.
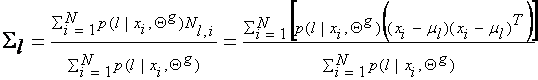
Summarizing, the
estimates of the new parameters in terms of the old parameters (guessed
parameters super-indexed by g, Qg =(a1g, …, aMg ; q1g, …, qMg)) are as follows:

Note that the above equations
(11) perform both the expectation step and the maximization step
simultaneously. The algorithm proceeds by using the newly derived parameters as
the guess for the next iteration.
Example 3: 1D Distribution Mixture-Model-Fitting using EM
These SOCR Activities demonstrate fitting a number of polynomial,
distribution and spectral models to data: http://wiki.stat.ucla.edu/socr/index.php/SOCR_EduMaterials_ModelerActivities.
For instance, suppose we have a collection of 100 observations. The
first 20 of these observations are included in the table below. A histogram of
these observations is also shown below.
|
1 |
-2.51002 |
|
2 |
-2.51002 |
|
3 |
-1.5060121 |
|
4 |
-1.5060121 |
|
5 |
-1.5060121 |
|
6 |
-1.5060121 |
|
7 |
-1.5060121 |
|
8 |
-1.5060121 |
|
9 |
-1.5060121 |
|
10 |
-1.5060121 |
|
11 |
-1.5060121 |
|
12 |
-1.5060121 |
|
13 |
-1.5060121 |
|
14 |
-1.5060121 |
|
15 |
-1.5060121 |
|
16 |
-0.502004 |
|
17 |
-0.502004 |
|
18 |
-0.502004 |
|
19 |
-0.502004 |
|
20 |
-0.502004 |
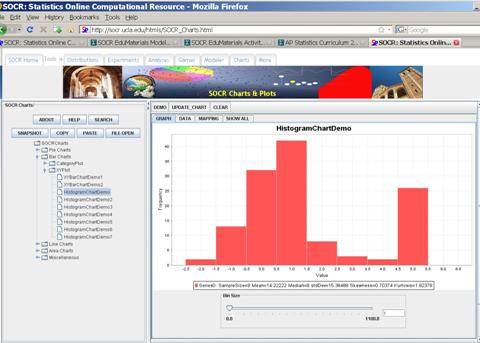
These data was generated using SOCR Modeler:
http://socr.ucla.edu/htmls/SOCR_Modeler.html
http://wiki.stat.ucla.edu/socr/index.php/SOCR_EduMaterials_Activities_RNG
And the Histogram was obtained using SOCR Charts:
http://socr.ucla.edu/htmls/SOCR_Charts.html
http://wiki.stat.ucla.edu/socr/index.php/SOCR_EduMaterials_Activities_Histogram_Graphs
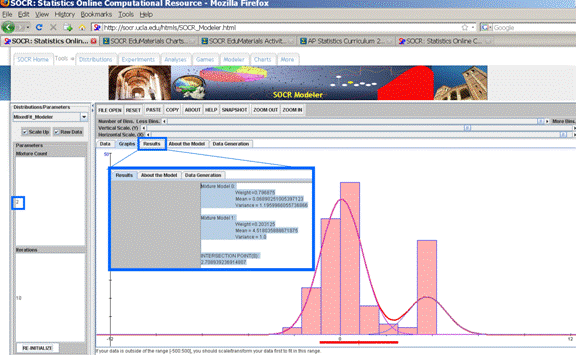
If these 100 random observations are copy-pasted in SOCR Modeler (http://socr.ucla.edu/htmls/SOCR_Modeler.html), we can fit in a mixture of 2-Normal Distributions to these data, as shown in the figure below. The quantitative results of this Em fit of 2 Normal distributions to these data is reported in the Results panel.
Mixture Model 0: Weight =0.796875 Mean =
0.06890251005397123 Variance =
1.1959966055736866 Mixture Model 1: Weight =0.203125 Mean = 4.518035888671875 Variance = 1.0 INTERSECTION
POINT(S): 2.708939236914807
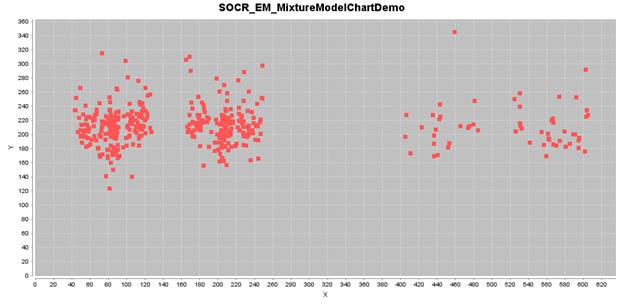
Example 4: 2D Point Clustering and Classification using EM
We can use the SOCR EM Chart, to enter real 2D data or simulate such
data. The figure below shows the plot of knee pain data (courtesy of Colin
Taylor, MD, TMT Medical).
·
http://socr.ucla.edu/htmls/SOCR_Charts.html
If we select four 2D Gaussian kernels, we can run iteratively the EM
mixture-modeling algorithm to estimate the 4-clusters and finally classify the
points in this knee-pain dataset, as shown in the figure below.
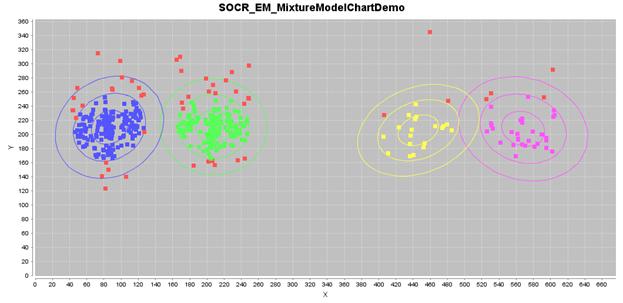
The estimated 2D Gaussian kernels are reported in the table below.
|
Kernel:1 |
Color[r=85,g=85,b=255] |
|
mX = 12.109004237402402 |
mY = 117.82567907801044 |
|
sXX = 21.032925241685124 |
sXY = 78.7581941291246 |
|
sYX = 78.7581941291246 |
sYY = 24.09825818460926 |
|
weight = 0.430083268505667 |
likelihood = -10.208282817360221 |
|
Kernel:2 |
Color[r=85,g=255,b=85] |
|
mX = 149.38436424342427 |
mY = 192.71749953677403 |
|
sXX = 20.76028543262703 |
sXY = -8.73565367620904 |
|
sYX = -8.73565367620904 |
sYY = 22.72742021102665 |
|
weight = 0.39741243015152933 |
likelihood = -10.208282817360221 |
|
Kernel:3 |
Color[r=255,g=255,b=85] |
|
mX = 384.0858049258344 |
mY = 130.33122378944105 |
|
sXX = 23.636423858007923 |
sXY = 135.0559608391195 |
|
sYX = 135.0559608391195 |
sYY = 21.654643444147702 |
|
weight = 0.06547362085865455 |
likelihood = -10.208282817360221 |
|
Kernel:4 |
Color[r=255,g=85,b=255] |
|
mX = 384.0858049258344 |
mY = 147.83898638544392 |
|
sXX = 25.080070954205347 |
sXY = -94.47319742216496 |
|
sYX = -94.47319742216496 |
sYY = 24.591327584325068 |
|
weight = 0.09143983449841454 |
likelihood = -10.208282817360221 |
Example 5: 3D Brain Tissue Classification using EM and Mixture Modeling
A demonstration of a 3D data analysis using the SOCR EM Mixture model is included in the LONI Viz Manual (http://www.loni.ucla.edu/download/LOVE/LOVE_User_Guide.pdf). This example shows how 3D brain imaging data may be segmented into three tissue types (White Matter, Gray Matter and Cerebra-spinal Fluid). This is achieved by LONI Viz (Dinov et al., 2006) sending the segmentation tasks to SOCR and SOCR returning back the 3D segmented volumes, which are superimposed dynamically on top of the initial anatomical brain imaging data in real time. The figure below illustrates this functionality. Other external computational tools could also invoke SOCR statistical computing resources directly by using the SOCR JAR binaries (http://www.socr.ucla.edu/htmls/SOCR_Download.html) and the SOCR Documentation (http://www.socr.ucla.edu/docs).
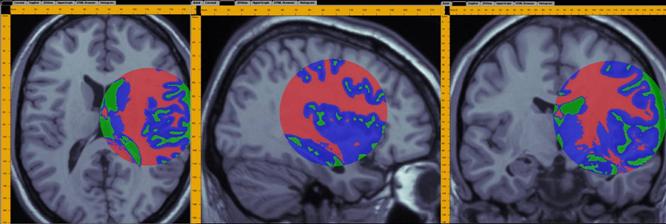
5. Online SOCR Demos
·
http://wiki.stat.ucla.edu/socr/index.php/AP_Statistics_Curriculum_2007_Estim_MOM_MLE
·
http://wiki.stat.ucla.edu/socr/index.php/SOCR_EduMaterials_ModelerActivities
·
http://socr.ucla.edu/htmls/SOCR_Modeler.html
·
http://wiki.stat.ucla.edu/socr/index.php/SOCR_EduMaterials_ModelerActivities_MixtureModel_1
6. References:
A.P.Dempster, N.M.
Laird, and D.B. Rubin. Maximum-likelihood from incomplete data via the EM
algorithm. J. Royal Statist. Soc. Ser. B., 39, 1977.
C. Bishop. Neural
Networks for Pattern Recognition. Clarendon Press,
J.A. Bilmes. A
Gentle Tutorial of the EM Algorithm and its Application to Parameter Estimation
for Gaussian Mixture and Hidden Markov Models. Department of Electrical
Engineering and Computer Science, U.C. Berkeley,
Dinov ID, Valentino D, Shin BC, Konstantinidis F, Hu G, MacKenzie-Graham A, Lee EF, Shattuck DW, Ma J, Schwartz C and Toga AW. LONI Visualization Environment, Journal of Digital Imaging. Vol. 19, No. 2, 148-158, June 2006.
Z. Ghahramami and
M. Jordan. Learning from incomplete data. Technical Report AI Lab Memo No.
1509, CBCL Paper No. 108, MIT AI Lab, August 1995.
M. Jordan and R.
Jacobs. Hierarchical mixtures of experts and the EM algorithm. Neural
Computation, 6:181–214, 1994.
M. Jordon and L.
Xu. Convergence results for the EM approach to mixtures of experts
architectures. Neural Networks, 8:1409–1431, 1996.
L. Rabiner and
B.-H. Juang. Fundamentals of Speech Recognition. Prentice Hall Signal
Processing Series, 1993.
R. Redner and H.
Walker. Mixture densities, maximum likelihood and the EM algorithm.
C.F.J. Wu. On the
convergence properties of the EM algorithm. The Annals of Statistics,
11(1):95–103, 1983.
L. Xu and M.I.
Jordan. On convergence properties of the EM algorithm for Gaussian mixtures. Neural
Computation, 8:129–151, 1996.